Numerical integration is straightforward to implement; however, “the curse of dimensionality” exists. This can be expressed mathematically as:
where:
: Represents the number of points.
: Indicates the product of from to .
: Shows that the number of grids is raised to the power of .
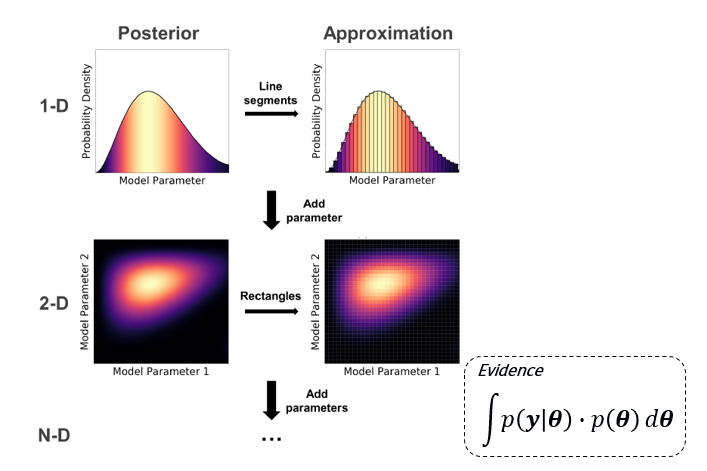
“A Conceptual Introduction to Markov Chain Monte Carlo Methods”, Joshua Speagle (2019)
Optional 💽
🖱️Right-click and select “Save link as” to download the exercise HERE 👈 so you can work on it locally on your computer
Hint
Start with formulating the likelihood function! Useful formulas for simple beams.
Prior GPa
The measurement error is obtained by the following formula:
For a given , the probability density of having a measurement = 50 is the probability density of the normal distribution with mean 0 and standard deviation 1, evaluated at
Imports¶
import matplotlib.pyplot as plt
import numpy as np
from scipy import statsProblem setup¶
# Fixed parameters
I = 1e9 # mm^4
L = 10_000 # mm
Q = 100 # kN
# Measurements
d_meas = 50 # mm
sigma_meas = 10 # mm
# Prior
E_mean = 60 # GPa
E_std = 20 # GPaForward model¶
def midspan_deflection(E):
return Q * L ** 3 / (48 * E * I)Bayesian functions¶
## Write your code where the three dots are
def likelihood(E):
return ...
def prior(E):
return ...Perform numerical integration¶
## Write your code where the three dots are
E_values = np.linspace(-20, 140, 1000)
# Calculate prior and likelihood values
prior_values = ...
likelihood_values = ...
evidence = ... # use the trapezoidal rule to integrate
posterior_values = ... Posterior summary¶
Hint
CDF:
Mean:
Standard deviation:
## Write your code where the three dots are
# Mean
mean = ... # use the trapezoidal rule to integrate
print(f'Mean: {mean:.2f} GPa')
# Median
cdf = np.cumsum(posterior_values) * np.diff(E_values, prepend=0)
median = E_values[np.argmin(np.abs(cdf - 0.5))]
print(f'Median: {median:.2f} GPa')
# Standard deviation
std = ... # use the trapezoidal rule to integrate
print(f'Standard deviation: {std:.2f} GPa')
# 5th and 95th percentiles
percentiles = np.interp([0.05, 0.95], cdf, E_values)
print(f'5th percentile: {percentiles[0]:.2f} GPa')
print(f'95th percentile: {percentiles[1]:.2f} GPa')Plot¶
plt.plot(E_values, prior_values, label='Prior')
plt.plot(E_values, likelihood_values, label='Likelihood')
plt.plot(E_values, posterior_values, label='Posterior')
plt.xlabel('E (GPa)')
plt.ylabel('Density')
plt.ylim([0, 0.045])
plt.legend()
plt.show()Complete Code! 📃💻
Here’s the complete code that you would run in your PC:
import matplotlib.pyplot as plt
import numpy as np
from scipy import stats
# Fixed parameters
I = 1e9 # mm^4
L = 10_000 # mm
Q = 100 # kN
# Measurements
d_meas = 50 # mm
sigma_meas = 10 # mm
# Prior
E_mean = 60 # GPa
E_std = 20 # GPa
#forward model
def midspan_deflection(E):
return Q * L ** 3 / (48 * E * I)
def likelihood(E):
return stats.norm.pdf(d_meas, loc=midspan_deflection(E), scale=sigma_meas)
def prior(E):
return stats.norm.pdf(E, loc=E_mean, scale=E_std)
#numerical integration
E_values = np.linspace(-20, 140, 1000)
prior_values = prior(E_values)
likelihood_values = likelihood(E_values)
evidence = np.trapz(prior_values * likelihood_values, E_values)
posterior_values = prior_values * likelihood_values / evidence
#posterior summary
# Mean
mean = np.trapz(E_values * posterior_values, E_values)
print(f'Mean: {mean:.2f} GPa')
# Median
cdf = np.cumsum(posterior_values) * np.diff(E_values, prepend=0)
median = E_values[np.argmin(np.abs(cdf - 0.5))]
print(f'Median: {median:.2f} GPa')
# Standard deviation
std = np.sqrt(np.trapz((E_values - mean) ** 2 * posterior_values, E_values))
print(f'Standard deviation: {std:.2f} GPa')
# 5th and 95th percentiles
percentiles = np.interp([0.05, 0.95], cdf, E_values)
print(f'5th percentile: {percentiles[0]:.2f} GPa')
print(f'95th percentile: {percentiles[1]:.2f} GPa')
plt.plot(E_values, prior_values, label='Prior')
plt.plot(E_values, likelihood_values, label='Likelihood')
plt.plot(E_values, posterior_values, label='Posterior')
plt.xlabel('E (GPa)')
plt.ylabel('Density')
plt.ylim([0, 0.045])
plt.legend()